Erdre
Quickstart
First, install Erdre and the requirements as specified here: Installation
To start using Erdre, follow the steps below.
1. Setup DVC
There are 2 options when setting up DVC:
Option A: Simple projects without need for tracking results
Initialize DVC with dvc init --no-scm.
The --no-scm option specifies that we do not want the DVC-related files to be
tracked by git, because this repository is agnostic to project-specific things
(data files, plots, metrics). This is recommended when there is no need to
track experiments in the repository, for simple projects, testing or developing
of the pipeline itself.
Option B: Track experiments using git
For long-term projects, it is recommended that you track DVC-related files with git, to take full advantage of the DVC functionality. Follow these steps:
- Fork this repository.
- Remove the lines under
# project-specific filesin .gitignore. - Initialize DVC with
dvc init.
2. Add data
To add your data to the pipeline, you have two options:
- Option A: Place the data files in the folder
assets/data/raw/. - Option B: If you want to easily swap between several data sets:
- Place the files in a subfolder
assets/data/raw/[NAME OF DATASET], where[NAME OF DATASET]is your chosen name of the data. - In
params.yaml, set the parameterdatasetto the name of your data set.
- Place the files in a subfolder
NB: Currently only .csv-files are supported.
Example with a data set called data01 (option B):
assets/
├── data/
| └── raw/
| ├── data01/
| | ├── experiment_01.csv
| | ├── experiment_02.csv
| | ├── experiment_03.csv
| | ├── ...
| | ├── experiment_17.csv
| | └── experiment_18.csv
| └─── README.md
├── metrics/
├── plots/
└── profiling/
The data set name is then specified in params.yaml:
profile:
dataset: data01
...
3. Specify parameters
In params.yaml, specify the name of the target variable (must match the name
of the column containing the target variable in the data files). Example with
target variable TargetVar:
clean:
# target: Name of target variable.
target: TargetVar
The rest of the parameters can also be adjusted. Documentation of the
parameters is found in
params.yaml.
4. Running experiments
Run experiments by executing this command on the main directory:
dvc repro
On some systems, dvc cannot be run as a standalone command, in which case you
will need to run:
python3 -m dvc repro
To run only parts of the pipeline, run:
dvc repro [STAGE NAME]
For example:
dvc repro profile # will only run the profiling stage
dvc repro featurize # will only run the stages up until and including the featurize stage
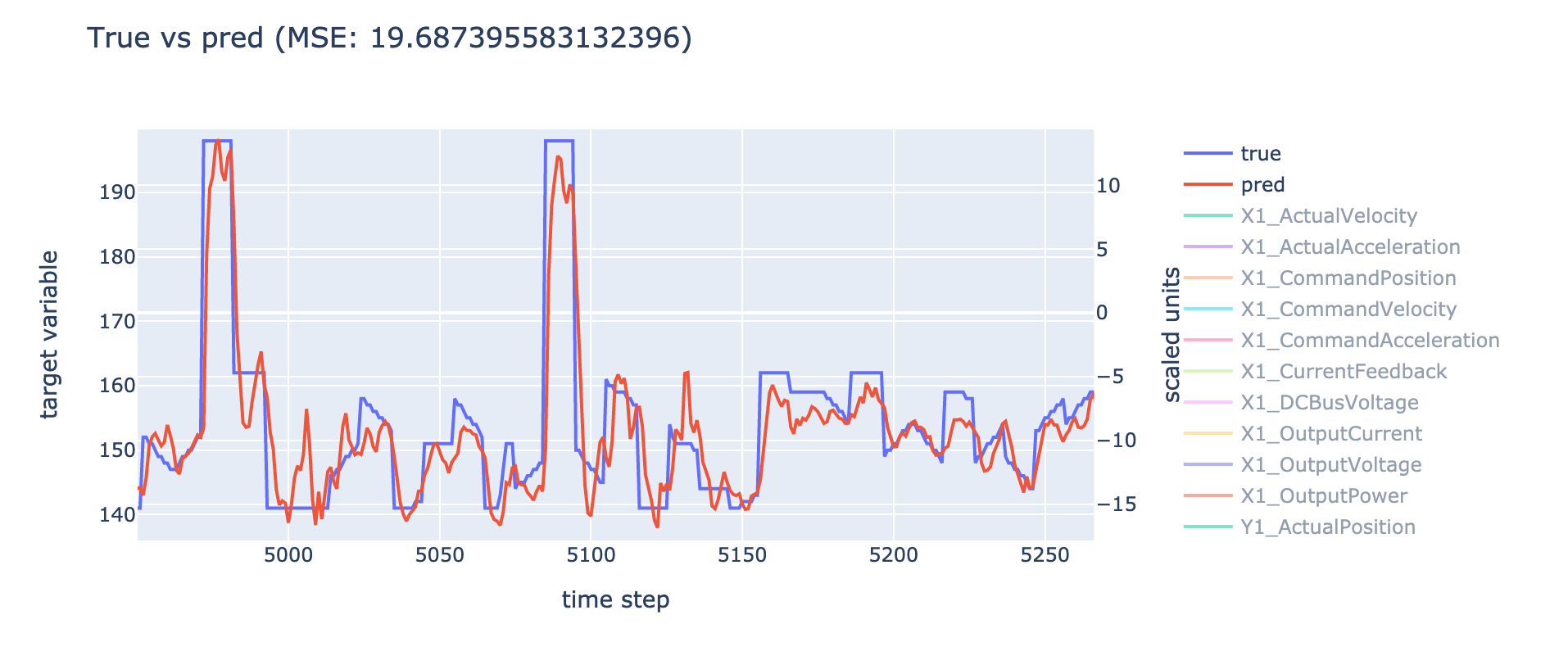
Example of results
When the trained model is evaluated, the program produces two plots that
visualizes the predictions on the test set. These plots are placed on the
folder assets/plots/, and show the following data:
prediction.html: The true target values compared to the predicted target values. Only the first value of each target sequence is used in the plot, and all of this values are connected and plotted as a line. The features used in the model are also plotted.
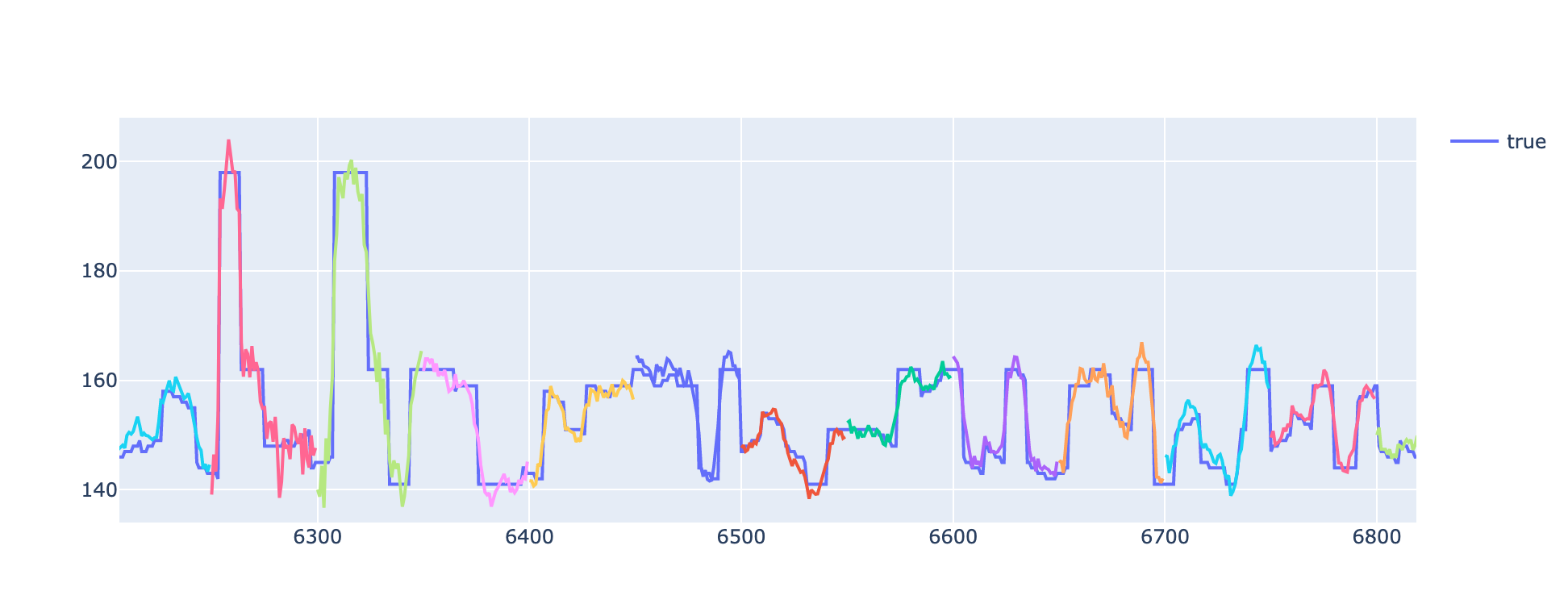
prediction_sequences.html: Individual predicted target sequences are plotted against the true values. The predicted sequences are shown in different colors to easily distinguish between them. Only a subset of the predicted target sequences are shown, in order to avoid overlapping and make the plot easier to interpret.
Next: Overview of pipeline